Python Bindings for Performance Optimization: A Zero to One Guide
Published:
This article describes techniques to accelerate a Python codebase by exposing parallelized C++ functions using PyBind. It then analyzes the results of the optimization in which parallelizing one 40-line function in a 700-line program yielded up to a 3X end-to-end speedup.
Introduction:
I accelerate an existing Python codebase while avoiding a full rewrite to keep the interface, visualization system, and data formats in tact. First I use cProfile to determine the function that consumes the most compute time. Next I implement a parallelized mathematical equivalent of the bottleneck function using concurrency facilities in modern C++ and achieve intra-process concurrency. I write a script to compile and test this function into a Python-importable module using PyBind. I then modify the original Python codebase to import and call the C++ function when requested from the command line. Lastly I measure an up to 3X performance improvement and provide observations.
Initial Execution:
The Python codebase we want to accelerate is an estimator for Gaussian Mixture Models (GMM). I’m interested most in the heart of the algorithm itself. The visualization system is useful for research purposes, but for production purposes it does not need to run. For that reason, I start by running the following command line which disables visualization and executes only the mathematical implementation of the estimator:
time python3 gmm_segmentation.py --first-image=example_data/church.jpg --components=3 --iterations=8 --visualization=0 --precompiled-num-threads=0
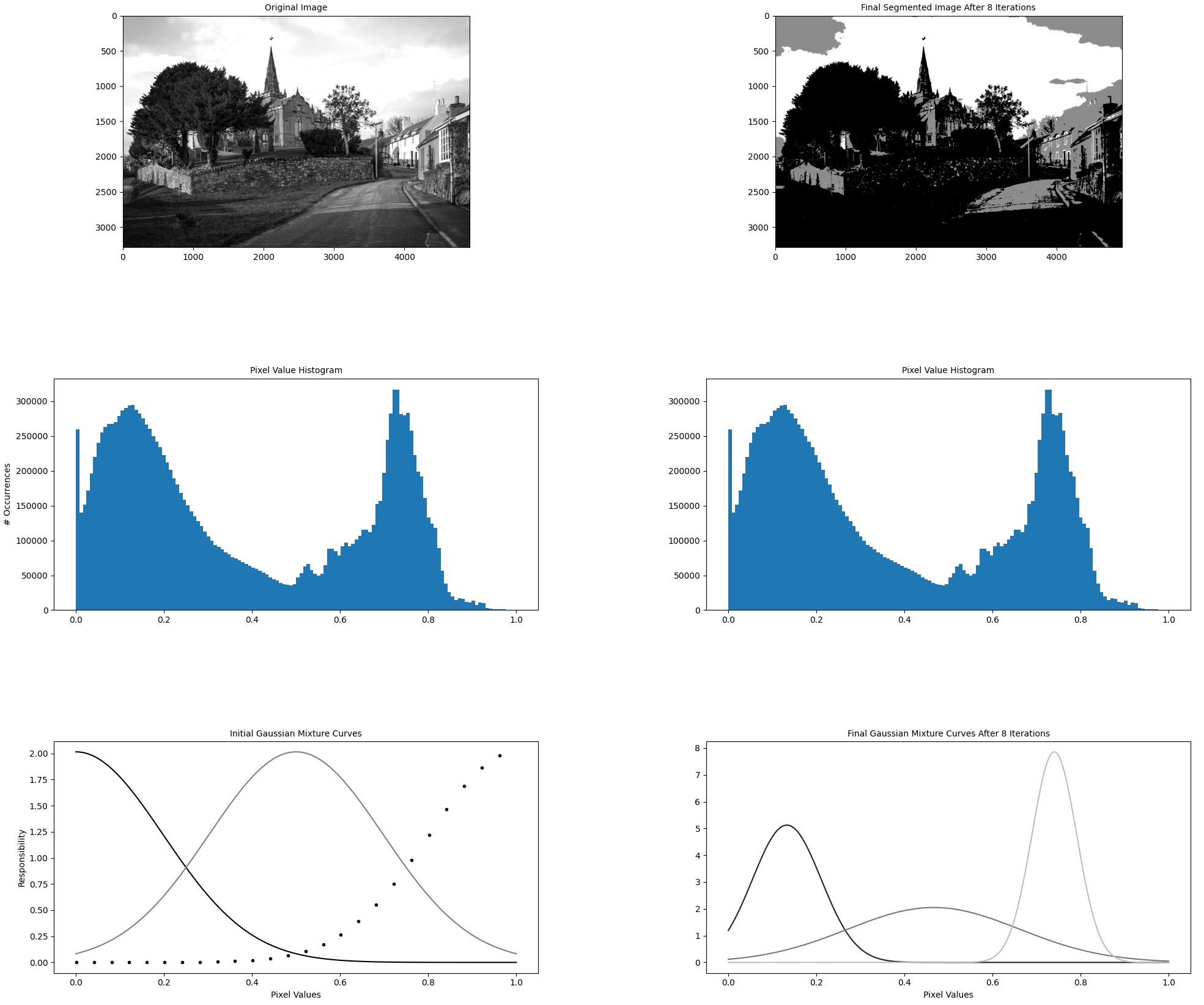
The driver function of the codebase is in the file gmm_segmentation.py which is where our efforts will go. --first-image specifies the location on disk that contains the image for which we want to compute a GMM. We will keep it constant. The algorithm hyperparameters --components and --iterations are hand-selected and not relevant for this article. While they do influence algorithm runtime (runtime increases as the number of components and iterations increases), we will keep them constant at all times. As discussed, --visualization toggles the visualization capability which outputs figures explaining the algorithm state as shown below. --precompiled-num-threads toggles the use of the accelerated C++ function. We will start with this parameter set to 0 meaning we run only the original Python implementation.
Observations upon Initial Profiling:
I insert cProfile calls inside the main() function of gmm_segmentation.py. The profiling capability is compact requiring only two lines to construct and enable the profiler before the business logic gets executed and 5 lines to summarize and display the results afterward. The function calls initialize_expectation_maximization() and execute_expectation_maximization() contain all of the logic we want to profile.
Upon executing the command line in the previous section on an AMD 3975WX CPU, we see the following (abridged) output:
ncalls tottime percall cumtime percall filename:lineno(function)
16 12.900 0.806 36.050 2.253 /home/a/gmm/gmm_segmentation.py:109(compute_expsum_stable)
241 8.690 0.036 8.690 0.036 {method 'reduce' of 'numpy.ufunc' objects}
48 5.702 0.119 5.702 0.119 /home/a/.local/lib/python3.10/site-packages/scipy/stats/_continuous_distns.py:289(_norm_pdf)
8 4.791 0.599 22.844 2.856 /home/a/gmm/gmm_segmentation.py:176(compute_expectation_responsibilities)
48 4.131 0.086 15.788 0.329 /home/a/.local/lib/python3.10/site-packages/scipy/stats/_distn_infrastructure.py:2060(pdf)
48 3.042 0.063 3.042 0.063 {built-in method numpy.core._multiarray_umath._insert}
1 2.623 2.623 44.719 44.719 /home/a/gmm/gmm_segmentation.py:209(execute_expectation_maximization)
real 0m45.572s
user 0m37.171s
sys 0m18.049s
The profiler recursively measures the time consumed by the functions that get called between the profiler.enable() and profiler.disable() calls. The functions are displayed in decreasing order of compute time consumed. Due to recursive operation, if functionA() internally calls functionB(), time spent in both functions is considered in the profiler ranking even though the profiler is only enabled and disabled once.
We see that the hottest function is compute_expsum_stable() in gmm_segmentation.py. Along with NumPy operators, we see also that compute_expsum_stable() calls a SciPy function, scipy/stats/_continuous_distns.py:289(_norm_pdf) (a.k.a scipy.stats.norm.pdf() in the actual code), which is listed as the third hottest. Therefore I decide to re-implement compute_expsum_stable() in C++ with hopes of increasing its performance.
Implementing, Compiling, and Importing a Custom C++ Module
NB: To illustrate this without the complexity of the GMM codebase, I made a separate and compact repo https://github.com/alexhagiopol/pybind_examples with no other purpose.
A multithreaded C++ equivalent of compute_expsum_stable() is implemented in computeExpsumStable() in PrecompiledFunctions.cpp. The re-implemented function is mathematically equivalent to the Python original and calls no external math functions other than basic matrix element access operators from the Eigen library. If using only a single thread, I expected the C++ function to underperform the Python version because the C++ function (a) naively re-implements SciPy’s scipy.stats.norm.pdf() without care for memory alignment or other lower level optimizations, (b) the original Python code uses vectorized syntax, and (c) the large 3D matrix P must be copied twice when calling the C++ function due to Eigen’s lack of official support for 3D matrices.
To compile the C++ code, the CMakeLists.txt file defines the build procedure for the Python module precompiled_functions which will land in a build folder precompiled_functions_build. The file re-uses PyBind’s own CMake module generation function. The Python script build_precompiled_functions.py automates the build process from build folder generation, to CMake invocation, to binaries generation. The Python module is imported by gmm_segmentation.py in the initialization of the GMM_Estimator object. The Python implementation of compute_expsum_stable() calls either a pure Python implementation or the imported C++ implementation depending on the user’s selection in the --precompiled-num-threads command line parameter. If set to 0, the pure Python implementation of compute_expsum_stable() will run. If set to 1 or more, that many threads will be given to the C++ implementation instead.
Observations upon Second Profiling:
With a C++ equivalent of the hottest function implemented, we give it one thread and do a second profiling run to compare against the pure Python version:
time python3 gmm_segmentation.py --first-image=example_data/church.jpg --components=3 --iterations=8 --visualization=0 --precompiled-num-threads=1
results in the following (abridged) profiler ranking:
ncalls tottime percall cumtime percall filename:lineno(function)
16 25.568 1.598 25.568 1.598 {built-in method precompiled_functions_build.precompiled_functions.computeExpsumStable}
8 4.878 0.610 19.853 2.482 /home/a/gmm/gmm_segmentation.py:176(compute_expectation_responsibilities)
16 4.394 0.275 29.962 1.873 /home/a/gmm/gmm_segmentation.py:109(compute_expsum_stable)
1 2.769 2.769 38.986 38.986 /home/a/gmm/gmm_segmentation.py:209(execute_expectation_maximization)
8 0.891 0.111 15.928 1.991 /home/a/gmm/gmm_segmentation.py:157(compute_log_likelihood_stable)
129 0.482 0.004 0.482 0.004 {method 'reduce' of 'numpy.ufunc' objects}
real 0m39.814s
user 0m34.286s
sys 0m15.183s
Some observations:
- Despite not using more than a single thread, calling the C++ function has reduced the total time to execute the program from 45.6s to 39.8s. Already this is a positive result considering the aforementioned disadvantages of the C++ implementation.
- In the profiler ranking, we notice that the function
scipy/stats/_continuous_distns.py:289(_norm_pdf)is no longer present as we expect. This is expected sincescipy.stats.norm.pdf()is no longer called. - The Python function
compute_expsum_stablehas been deranked and replaced by the C++ functionprecompiled_functions.computeExpsumStablewhich is also expected because the bulk of the work is now done in the C++ implementation. - The number of
{method 'reduce' of 'numpy.ufunc' objects}has been reduced from 241 to 129 and with a time cost reduced from 8.7s to 0.48s. This is also expected: the number of numpy operations required is reduced because the math is now implemented in C++.
Observations upon Third Profiling:
Let’s see what happens when we let the 32-core & 64-thread AMD 3975WX CPU stretch its legs and use more than one thread for the C++ function. We’ll look only at total program runtime in this section.
- Single Python thread: 45.6s
- Single Python thread calling single-thread C++ function: 39.8s
- Single Python thread calling 4-thread C++ function: 21.4s
- Single Python thread calling 8-thread C++ function: 18.3s
- Single Python thread calling 8-thread C++ function: 15.9s
- Single Python thread calling 32-thread C++ function: 15.8s
- Single Python thread calling 64-thread C++ function: 15.5s
The results are quite encouraging: by parallelizing only a single function with intra-process concurrency, we’ve yielded a close to 3X acceleration of the entire program. Looking at the profiler ranking for the 64-thread run, we see that the C++ function is so fast that’s it’s no longer top-ranked indicating that we would want to optimize other parts of the system next:
ncalls tottime percall cumtime percall filename:lineno(function)
8 4.881 0.610 8.033 1.004 /home/a/gmm/gmm_segmentation.py:176(compute_expectation_responsibilities)
16 4.093 0.256 6.237 0.390 /home/a/gmm/gmm_segmentation.py:109(compute_expsum_stable)
1 2.562 2.562 15.041 15.041 /home/a/gmm/gmm_segmentation.py:209(execute_expectation_maximization)
16 2.143 0.134 2.143 0.134 {built-in method precompiled_functions_build.precompiled_functions.computeExpsumStable}
8 0.867 0.108 4.001 0.500 /home/a/gmm/gmm_segmentation.py:157(compute_log_likelihood_stable)
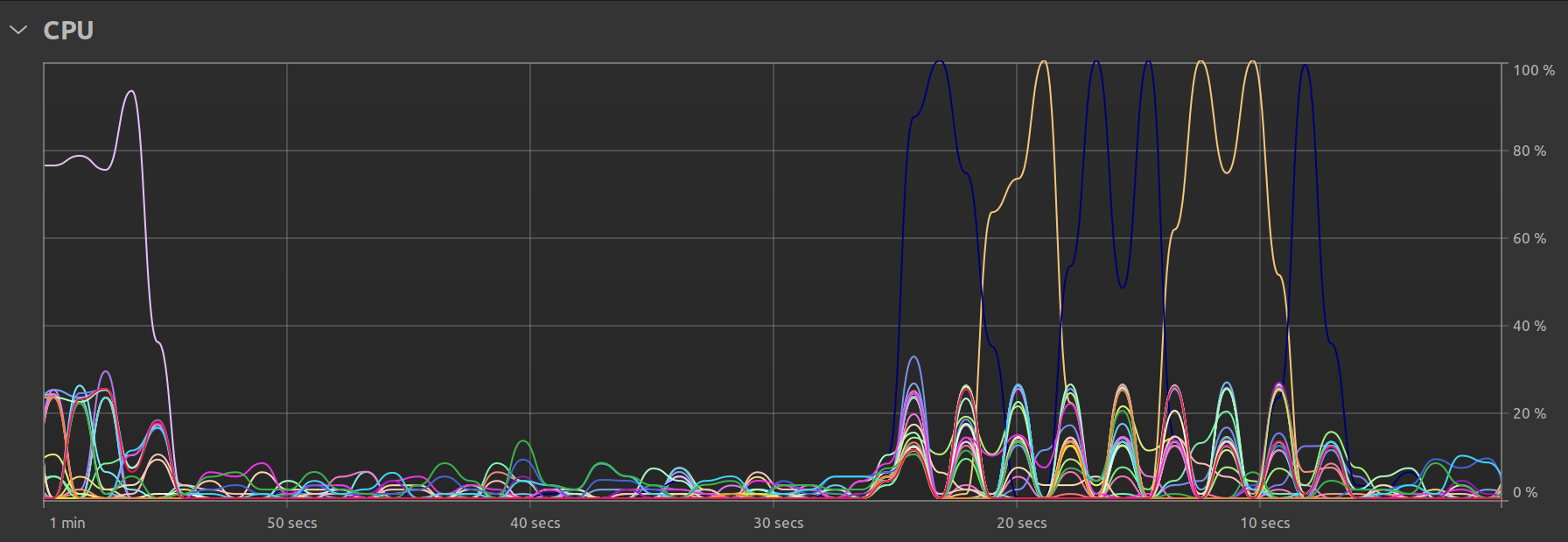
One final observation is my Ubuntu system’s own resource utilization graph. In the single threaded case, we clearly see that there is some parallel activity at program startup, but the bulk of the time is spent with just one thread doing all the work:
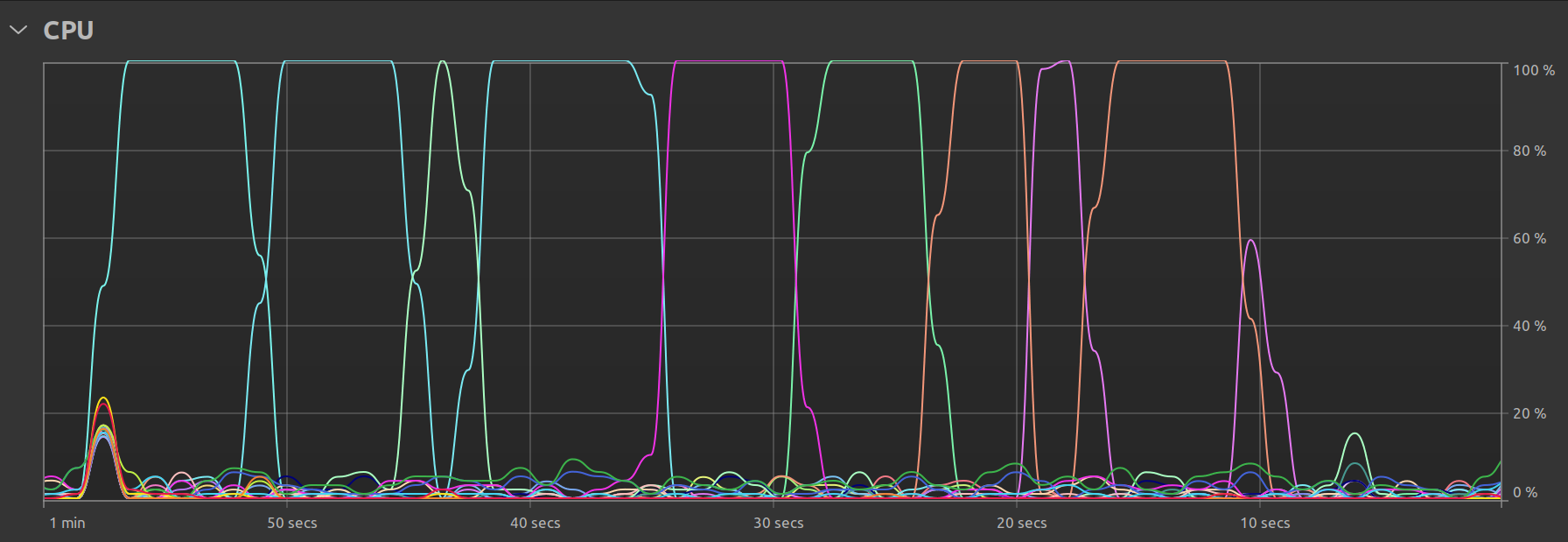
In the multi-threaded case, we can see that there is still one main thread but that there exist smaller “peaks” representing the times when the remaining available threads are called upon to perform the work: